Research
Over the past decade, technological advancements in next-generation sequencing and genomics have revolutionized the field of biology. Unfortunately, the unprecedented pace and volume of data being generated often exceeds the infrastructure and bioinformatic capabilities of many researchers. As a result, many disciplines have developed a divide between those actively involved in experimental research and data collection at the organismal level and those with the computational and bioinformatic skills to process these data. As a quantitative organismal biologist, my research continuously bridges this divide by integrating skills and training in computational genomics with experimental research in organismal biology while fostering collaborations with scientists on both sides. In particular, my research program utilizes these quantitative approaches to investigate the measurement, maintenance, and restoration of biodiversity (Fig. 1, below).
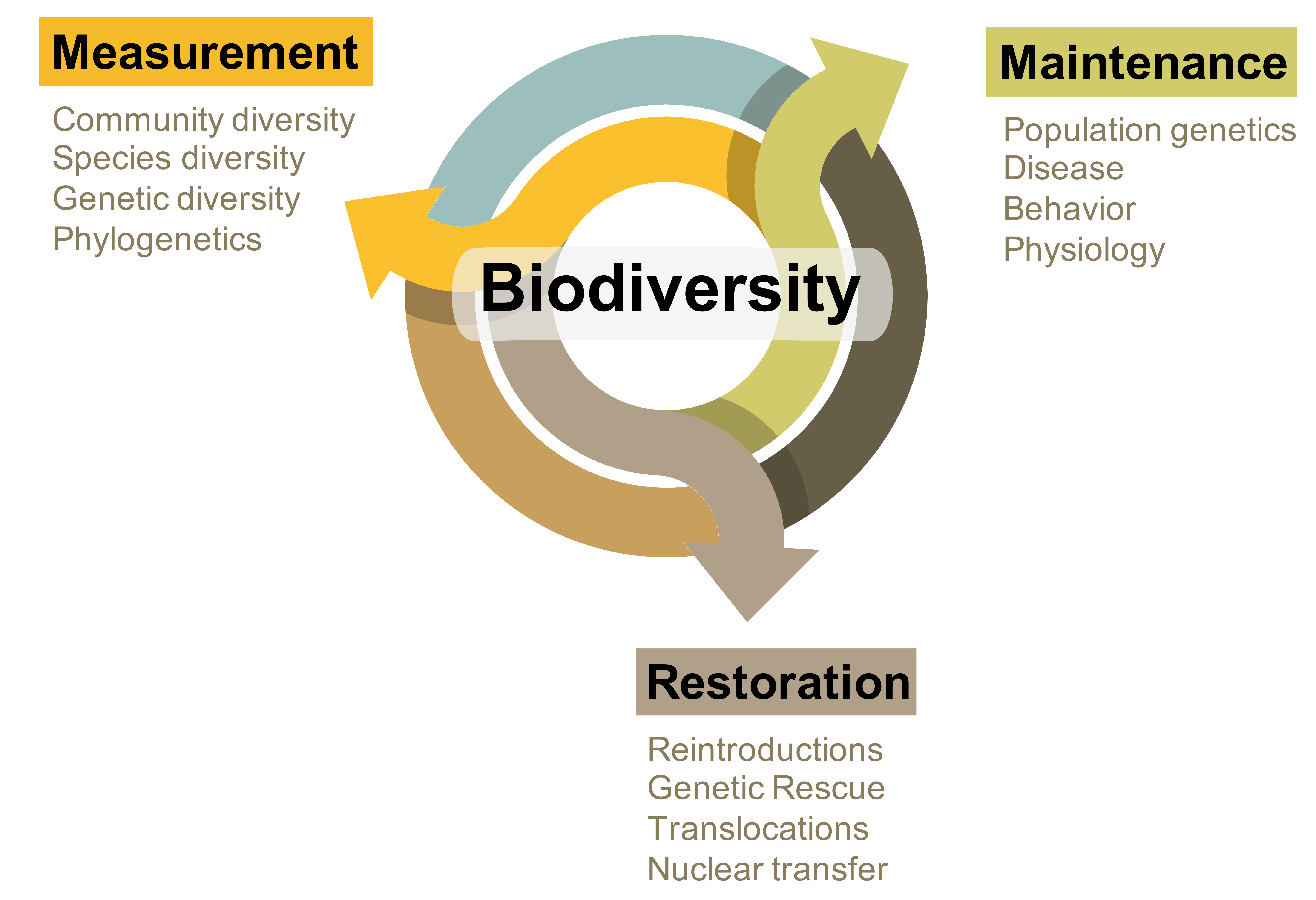
In one way or another, my lab’s various projects and interests intersect the chart in Fig. 1 (right), whether it be through the importance of understanding levels of genetic diversity, basic biology (e.g., physiology and behavior), or in directly translational results (e.g., population reintroductions). Furthermore, this paradigm is not limited to wild species, as many of the same concepts are directly applicable to domestic or otherwise agriculturally and economically important species. Below you will find specific examples of ongoing research projects in the lab.
Projects
-
Magnetoreception in Animals
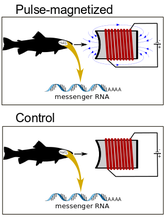
Over the last several decades, behavior experiments have shown that a variety of animals are capable of sensing Earth's magnetic field. Although several theories exist to explain this phenomenon, the underlying molecular basis remains unknown. Our research at UCF is trying to identify the possible location of a 'magnetoreceptor' and putative genes involved in a magnetic sense. We utilize various RNA-seq experiments in fish (i.e. salmonids) to quantify changes in gene expression after exposure to magnetic stimuli. We have examined both the brain and retina to date. There is essentially no change in gene expression in the retina, but numerous changes were observed in the brain. The changes in the brain have yielded 'candidate magnetoreception genes', such as the protein ferritin, in which ongoing experiments are examining more closely.
Funding: Air Force Office of Scientific Research (through Sönke Johnsen at Duke University)
Collaborators: Sönke Johnsen (Duke), Jay Wheeler (Duke), Ken Lohmann (UNC), Dave Ernst (UNC/UArk), Lewis Naisbett-Jones (UNC)
Assignees: TBD
-
Geomagnetic Biogeography
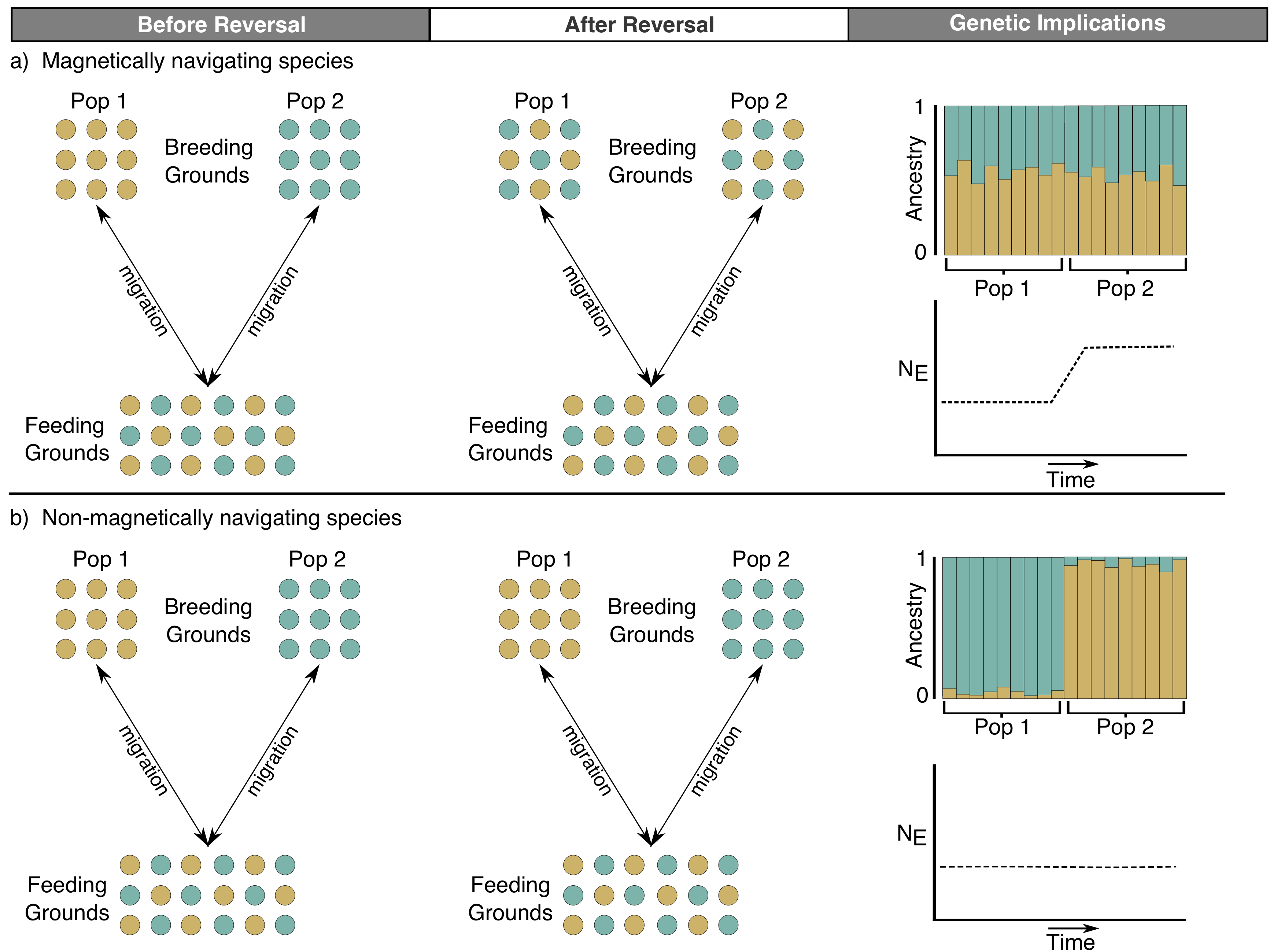
Earth’s magnetic field is quite dynamic - varying in strength, orientation, and even occasionally reversing polarity (swapping the North and South poles). It is possible this variation disrupts the navigation of species that depend on Earth’s magnetic field to migrate. Currently, we are outlining a conceptual approach for how population genetics can reveal changes in the demographic history and/or biogeography of species as a result of this variation - a phenomenon we call 'geomagnetic biogeography'.
We have provided some preliminary evidence in green sea turtles, where we recently showed that changes in gene flow between populations can account, at least partially, for a sudden increase in historical effective population size around the time of the last polarity reversal (780,000 years ago). This result, along with some great work by others, suggest that geomagnetic variation may have important consequences on the migration, and in turn, biogeography, of species that navigate using the geomagnetic field. Our goal is to continue work in this area in other 'magnetically sensitive' or navigating species to determine the role geomagnetic history plays in shaping species’ demography.Funding: N/A
Collaborators: Sönke Johnsen (Duke), Jay Wheeler (Duke), Roger Brothers (UNC), Ken Lohmann (UNC)
Assignees: TBD
-
American Shad Genome Sequencing

The American shad (Alosa sapidissima) genome: understanding the evolutionary novelties of the “fish that fed the Nation’s founders"
American shad are anadromous fish distributed throughout the North American Atlantic coast, and prior to the 20th century, were one of the most economically important fisheries in the U.S. Adults spend most of their life at sea, and return in the spring to their natal streams to spawn. This migratory behavior, like that of salmonids, is of particular interest to sensory biologists as the exact environmental cues employed to navigate to their natal areas remains unclear. It has been demonstrated in salmonids that the geomagnetic field can be used to navigate, but whether or not shad utilize the same method is unknown. American shad also have evolved several other unique features of interest to sensory biologists. First, unlike most fish, shad lack a distinct lateral line (a critical electrophysiological and mechanosensory ‘organ’). Second, shad can hear ultrasonic sounds up to 180 kHz (compared with 20 kHz in humans), one of the highest frequency ranges of any organism ever recorded and speculated to be a defense against cetacean echolocation (~40 – 150 kHz). Despite the unique sensory features of American shad, little research has focused on this species. One reason is the lack of genetic and genomic resources available. Not only will genomic resources facilitate evolutionary and functional studies of these unique sensory capabilities, but provide additional genetic resources to the wild fisheries managers to benefit the conservation and management of this important local resource. Currently, through a grant from Duke's Center for Genomics and Computational Biology, we are sequencing and assembling the first American shad genome.Funding: Duke Center for Genomic and Computational Biology
Collaborators: Sönke Johnsen (Duke), Jaw Wheeler (Duke)
Assignees: TBD